Figure 3
The advances and challenges of Gene Therapy for Duchenne Muscular Dystrophy
Jacques P Tremblay* and Jean-Paul Iyombe-Engembe
Published: 25 July, 2017 | Volume 1 - Issue 1 | Pages: 019-036
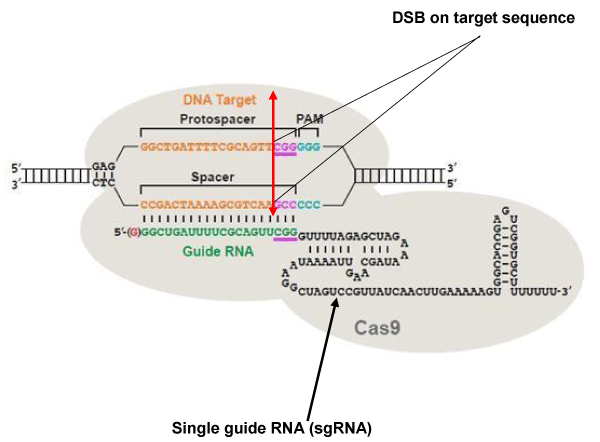
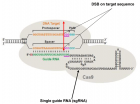
Figure 3:
The CRISPR / Cas9 system. The DNA target sequence (orange) is illustrated with the two strands separated. The sense strand of target sequence id followed at its 3’ end by the protospacer adjacent motif (PAM), which is NGG (in blue) for the SpCas9 nuclease. The cleavage site (double arrow in red) is located at 3 nucleotides (CGG in mauve) from the PAM towards the 5 ‘end. The variable portion of sgRNA (20 nucleotides in green) recognizes the target sequence by the Watson-Crick base pairing. The nuclease Cas9 (in light gray) is recruited at the PAM. The invariable part of sgRNA (nucleotides in black) stabilizes the whole structure. Image modified from [107].
Read Full Article HTML DOI: 10.29328/journal.jgmgt.1001003 Cite this Article Read Full Article PDF
More Images
Similar Articles
Recently Viewed
-
Intersecting Pathways: Examining Hildegard Peplau's and Rosemarie Parse's Nursing Theories through a Comparative LensMehtab Lalwani*, Rafat Jan and Salma Rattani. Intersecting Pathways: Examining Hildegard Peplau's and Rosemarie Parse's Nursing Theories through a Comparative Lens. Clin J Nurs Care Pract. 2023: doi: 10.29328/journal.cjncp.1001046; 7: 009-014
-
The Inverse Relationship between Acute Myocardial Infarction and Dissolved Oxygen Levels in WaterArturo Solís Herrera*,María del Carmen Arias Esparza,Ruth Isabel Solís Arias. The Inverse Relationship between Acute Myocardial Infarction and Dissolved Oxygen Levels in Water. J Nov Physiother Rehabil. 2025: doi: 10.29328/journal.jnpr.1001066; 9: 013-023
-
Screening of antibiotic residue in poultry in Kathmandu valley of Nepal: A cross-sectional studyRajiv Sapkota*,Ramila Raut,Sujan Khanal,Muna Gyawali,Dipesh Sahi. Screening of antibiotic residue in poultry in Kathmandu valley of Nepal: A cross-sectional study. Arch Pharm Pharma Sci. 2019: doi: 10.29328/journal.apps.1001017; 3: 079-081
-
Investigation of Stain Patterns from Diverse Blood Samples on Various SurfacesSonia Rajkumari*. Investigation of Stain Patterns from Diverse Blood Samples on Various Surfaces. J Forensic Sci Res. 2024: doi: 10.29328/journal.jfsr.1001061; 8: 028034
-
Needs in siblings of individuals with Down Syndrome and levels of coping Cali, ColombiaLina Fernanda Martínez Cadavid,Diana Marcela Gutiérrez Acosta,Luis Alexander Lovera Montilla*. Needs in siblings of individuals with Down Syndrome and levels of coping Cali, Colombia. J Adv Pediatr Child Health. 2021: doi: 10.29328/journal.japch.1001041; 4: 093-100
Most Viewed
-
Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth EnhancersH Pérez-Aguilar*, M Lacruz-Asaro, F Arán-Ais. Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth Enhancers. J Plant Sci Phytopathol. 2023 doi: 10.29328/journal.jpsp.1001104; 7: 042-047
-
Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case PresentationJulian A Purrinos*, Ramzi Younis. Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case Presentation. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001099; 8: 075-077
-
Feasibility study of magnetic sensing for detecting single-neuron action potentialsDenis Tonini,Kai Wu,Renata Saha,Jian-Ping Wang*. Feasibility study of magnetic sensing for detecting single-neuron action potentials. Ann Biomed Sci Eng. 2022 doi: 10.29328/journal.abse.1001018; 6: 019-029
-
Pediatric Dysgerminoma: Unveiling a Rare Ovarian TumorFaten Limaiem*, Khalil Saffar, Ahmed Halouani. Pediatric Dysgerminoma: Unveiling a Rare Ovarian Tumor. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001087; 8: 010-013
-
Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative reviewKhashayar Maroufi*. Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative review. J Sports Med Ther. 2021 doi: 10.29328/journal.jsmt.1001051; 6: 001-007

HSPI: We're glad you're here. Please click "create a new Query" if you are a new visitor to our website and need further information from us.
If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."